We are thrilled to announce that our collaborative research paper, "Multi-Institutional PET/CT Image Segmentation Using Federated Deep Transformer Learning", has been accepted for publication in the Elsevier journal, Computer Methods and Programs in Biomedicine.
The product of a unique collaborative effort between our group and esteemed partners at the Geneva University Hospital (HUG), RWTH Aachen University Hospital, and Imperial College London, the paper explores various approaches to PET/CT image segmentation in HN Cancer patients.
Key contributions and findings of the research include:
- The successful integration of purely attention-based transformers and Federated Learning (FL) algorithms for PET/CT Image segmentation.
- The application of the FL framework for PET/CT image segmentation, leading to a more generalizable model development in multi-center settings.
- The implementation of different FL frameworks, each designed to address specific challenges of a FL model, such as different learning paradigms, aggregation, robustness, privacy, and communication efficiency.
- A comprehensive comparison between center-based, centralized, and FL frameworks, highlighting their respective strengths and areas of improvement.
- An in-depth quantitative analysis performed on PET images, helping drive clinical evaluation of segmentation algorithms.
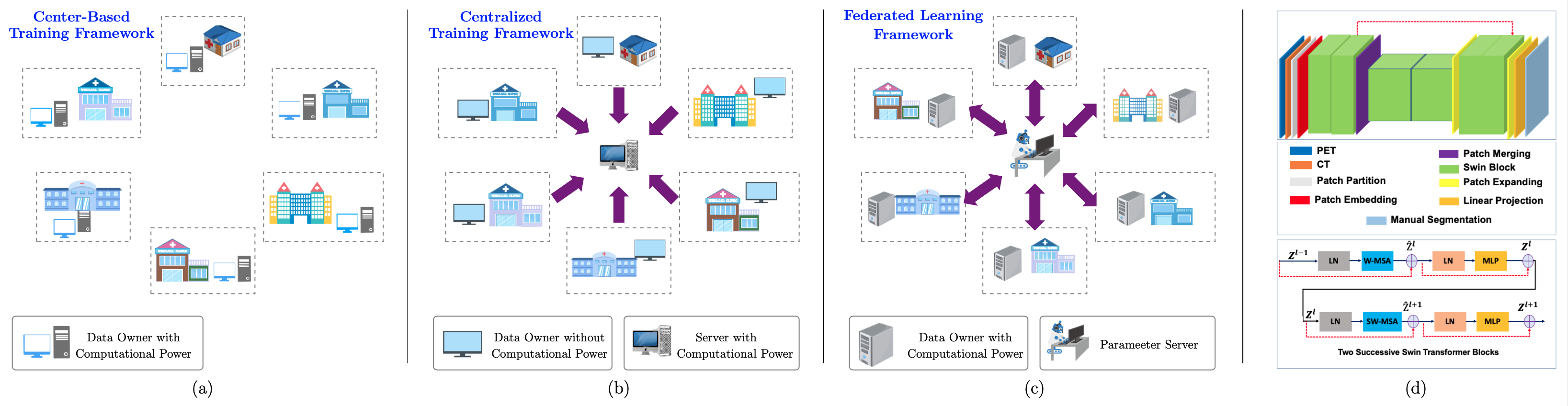
Visual illustration of (a) center-based training, (b): centralized training, (c): federated learning frameworks; (d) our network architecture.

3D views of PET/CT segmentation obtained from manual (red) and different algorithms on representative patients from different centers. (1-6 from left to right): center-based (CeBa), centralized (CeZe), clipping with the quantile estimator (ClQu), zeroing with the quantile estimator (ZeQu), federated averaging (FedAvg), lossy compression(LoCo), robust aggregation (RoAg), secure aggregation (SeAg), Gaussian differentially private federated averaging with adaptive quantile clipping (GDP-AQuCl).
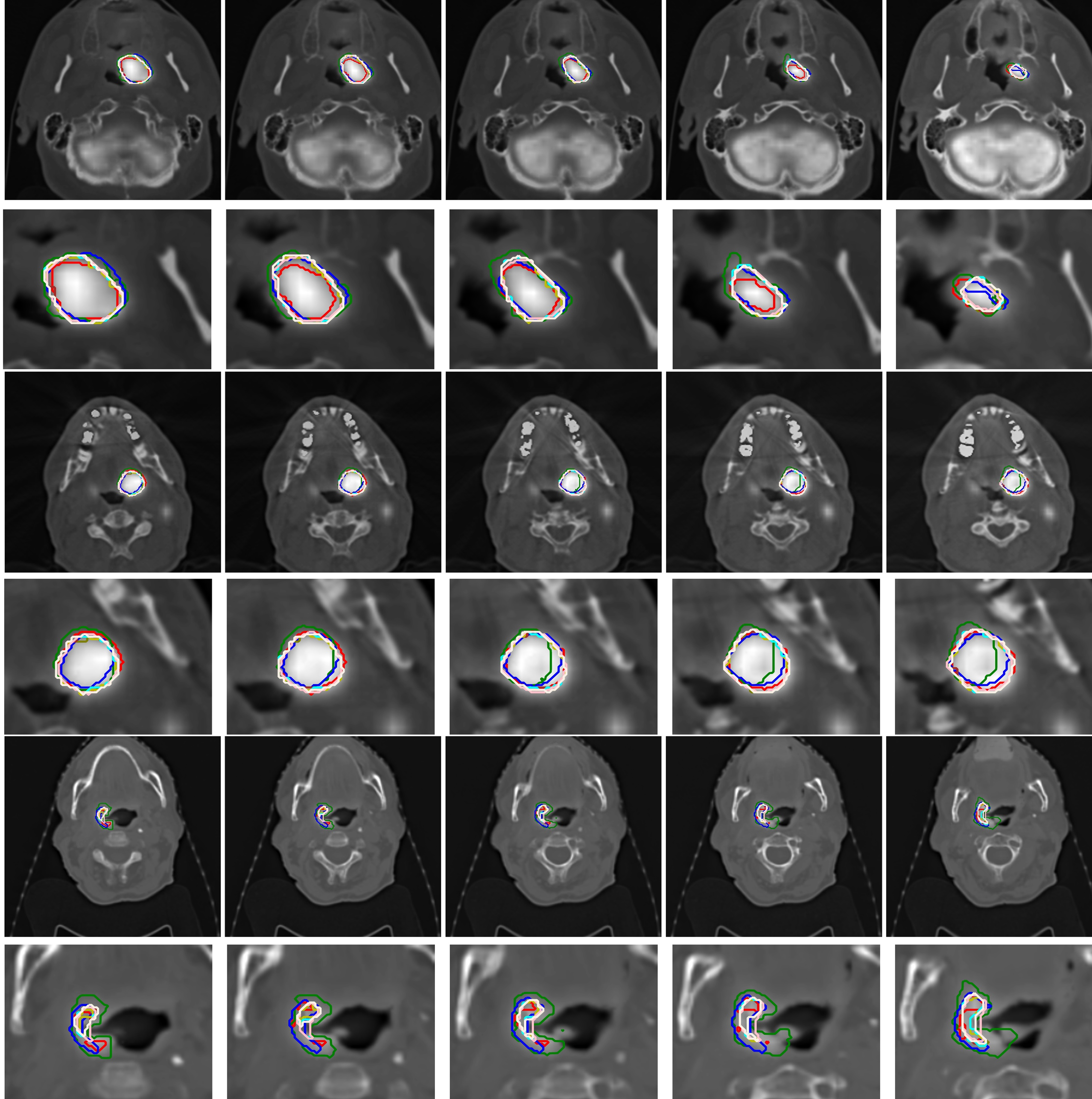
2D views of PET/CT segmentations obtained manually in three different cases: Red. CeBa: Green, CeZe: blue, ClQu: Brown, ZeQu: Olive, FedAvg: Orange, LoCo: Cyan, RoAg: Pink, SeAg: Linen, GDP-AQuCl: Yellow.



